2023
Welcome Sergei Rudnizky (postdoc, shared Taekjip Ha lab from 2022) and Sonia Weiner (undergraduate student).
Welcome summer students Kemi Abiodun, Kelly Chen, Annie Huang, Sean Kao, Chloe Liang, Kayra Ozturan, Sonia Weiner.
Congratulations to Dr. Ashlee Xinyu Feng, graduate of the CMDB program and recipient of the Biology Department Roseman Award for outstanding research in biochemistry!
Congratulations to Xiaona and Taibo for the collaborative paper with Mike Levine’s lab on GAF-dependent chromatin looping!
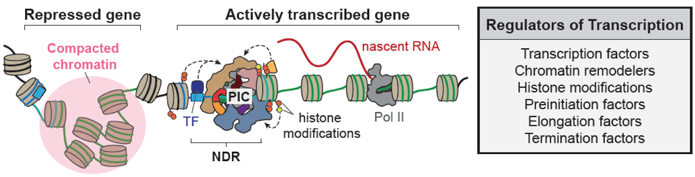
Our lab receives a 5-year MIRA grant award from NIGMS to investigate “Kinetic Mechanisms of Chromatin Remodeling and Transcription.” Congratulations to everyone who made it possible. Awesome effort.
Welcome Muhammad Bennani, Maggie Wang, Sophie Yao, Chloe Liang to the Spring Semester Advanced Cell Biology Lab Course in Single-Molecule Imaging.
Alice Chen joins the lab as a graduate student from the BCMB program. Welcome Alice!
2022
Congratulations to Dr. Jee Min Kim, graduate of the CMDB program and recipient of the Biology Department Roseman Award for outstanding research in biochemistry! All the best for a successful postdoc at NIH!
Congratulations to Xiaona Tang et al. on the publication of the NSMB paper on live imaging of GAGA factor! Great piece of work.
Big congratulations to Assistant Professor Vu Nguyen, new faculty member at University of California, San Diego. We will miss you!
Giho Park returns to the lab as an MD-PhD student, to continue CryoEM with Robert. Welcome back Giho!
Maryam Yamadi joins the lab as a new graduate student. Welcome Maryam!
August, 2021
Congratulation to Vu and Jee Min for their respective publications in Molecular Cell and eLife, and the collaborative venture with Francois Robert’s lab, also out in Molecular Cell.
July, 2021
Zelin Wei joins the lab as our new graduate student. Welcome!
June, 2021
Farewell to Ejlal and Giho, our gap year students on their way to MD-PhDs
December, 2020
Vu Nguyen submits his magnum opus to bioRxiv “Spatio-Temporal Coordination of Transcription Preinitiation Complex Assembly in Live Cells.” Way to go, Vu!
November, 2020
We are most pleased to be part of the NIH 4D nucleome program. Thanks to Xiaona Tang, lab lead for the funded Ha-Johnson-Wu consortium U01 proposal “Chromatin Function During Transcription and DNA Repair at Single-Molecule Resolution in Living Cells.”
August, 2020
Congratulations to Taibo for passing his oral exams!
July, 2020
We have a lab website on-line! Thanks to Jenna Louder.
A warm welcome to Nathan Jones, Ph.D. Biophysics, joining us from Ohio State!
June, 2020
Phase I reopening after a 3-month shut-down, and we are relieved to get back to work, cautiously.
May, 2020
Congratulations to Sun-Jay on his graduation from the Computer Science program, and many thanks for contributions to Sojourner, our single-molecule computation pipeline!
Congratulations to Eve (Ejlal) on her graduation from the MCB program and continuing as research technologist for her gap year!
Taibo Li, MD. Ph.D. student joins the lab, co-mentored with Prof. Alexis Battle, Dept. of Biomedical Engineering. Welcome!
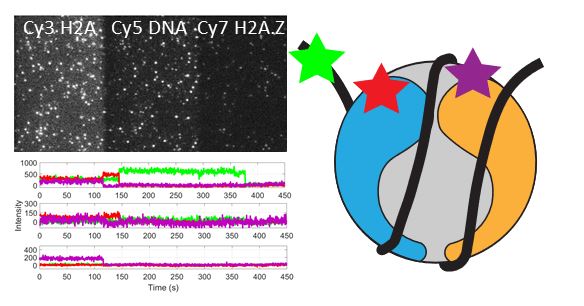
Congratulations to Anand and the team for their eLIFE publication “Live-cell single particle imaging reveals the role of RNA polymerase II in histone H2A.Z eviction.” A milestone for the lab’s entry into single-molecule imaging.
April 2020
Congratulations to Hin for his Croucher Foundation post-doctoral fellowship award!
March, 2020
The lab and Hopkins are shut down due to COVID19.
December, 2019
Welcome Hin Ling, PhD. Biochemistry, to the lab. From Hong Kong to snowy Baltimore!
July 2019
Congratulations to Robert for his NRSA post-doctoral fellowship award!
June, 2019
Giho Park graduates from the MCB program and re-joins the lab as research technologist.
June, 2019
Congratulations to Vivian, Pat, and Kai-Yu our undergrad researchers on their graduation from the MCB program at Hopkins!
January and April, 2019
Sheng and Qin, respectively, return to Canada, on the way to new positions in China. Many thanks for essential roles in setting up our Hopkins operation and our very best wishes!
July 2018
Congratulations to Matt for his NRSA post-doctoral fellowship award!
June, 2018
Ashlee Feng joins the lab as our second graduate student from CMDB, jointly mentored by TJ Ha.
March, 2018
Robert Louder, Ph.D. structural biology, arrives to his native Maryland from sunny Berkeley.
June, 2017
Xiaona Tang, Ph.D. Molecular Genetics, moves with Drosophila strains from our previous NIH lab to continue her postdoc at Hopkins.
April, 2017
Matt Poyton, Ph.D. Chemistry, joins the lab, a joint postdoc with TJ Ha. Welcome!
June, 2017
Jee-Min Kim joins the lab as our very first graduate student, from the CMDB program!
January, 2017
Gaku, Anand, and Vu from HHMI-Janelia join Carl at Homewood.
September, 2016
Sheng Liu, Ph.D. Molecular Genetics, and Qin Liu, B.Sc. join the lab from Vancouver, Canada.
July, 2016
Carl moves from HHMI Janelia’s Transcription Imaging Consortium to start the new lab at the Hopkins Homewood campus, Baltimore.